ete-view
visualization and rendering of phylogenetic trees and multiple sequence alignments.
Overview
ete-view allows to visualize or render tree images directly from the command line
Examples
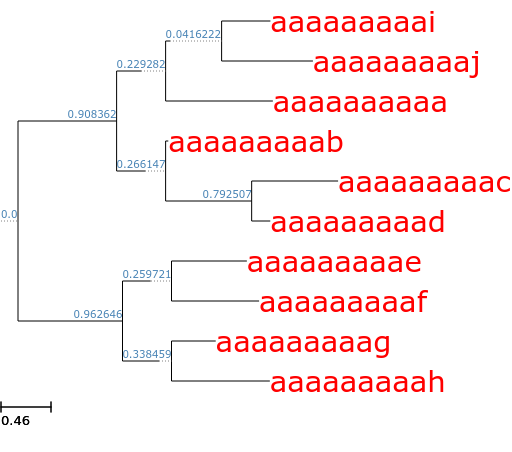
Basic visualization
ete-view allows to visualize one or more trees at a time
ete3 generate --random_branches -n 2 | ete3 view
Customization
ete-view allows a lot of customization, i.e. adding node faces
ete3 generate --random_branches | ete3 view --face 'value:@name, color:red, size:20, pos:b-right' --face 'value:@dist, color:steelblue, size:7, pos:b-top, nodetype:internal'
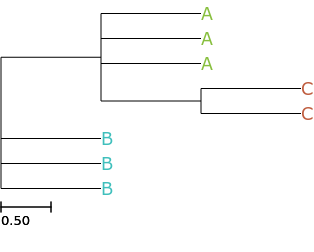
you can also use it to auto distribute colors in branches based on annotation
# auto distribute colors according to attributes ete3 view -t '((A, A, A, (C, C)), B, B, B);' --face 'value:@name, color:auto()'
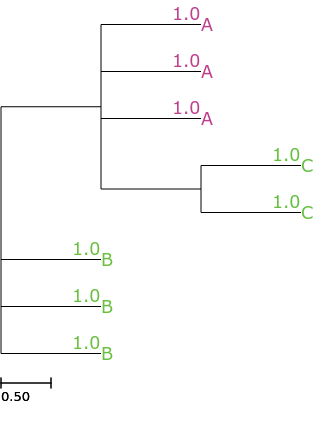
even using other attributes as a source
ete3 view -t '((A, A, A, (C, C)), B, B, B);' --face 'value:@name, color:auto()' --face 'value:@dist, pos:b-top, color:auto(@name)'
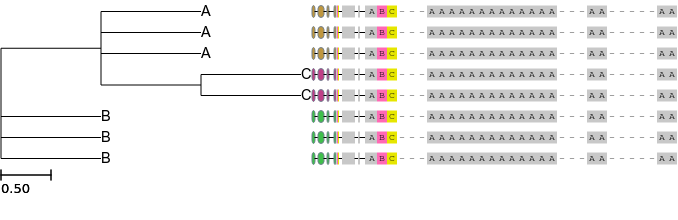
Visualizing tree and alignments
Common visualization templates are supported out of the box. i.e. alignments
ete3 view -t '(A, (B, C));' --alg alignment.fa
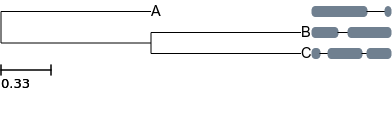
Although much more advanced customization is always possible
ete3 view -t '((A, A, A, (C, C)), B, B, B);' --face 'value:ABC---AAAAAA---AA-----AA, ftype:blockseq, bgcolor:auto(@name), pos:aligned' --face 'value:ABC---AAAAAAAAAAAAA---AA-----AA, ftype:compactseq, pos:aligned' --face 'value:ABC---AAAAAAAAAAAAA---AA-----AA, ftype:fullseq, pos:aligned'